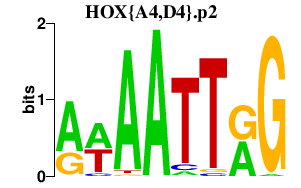
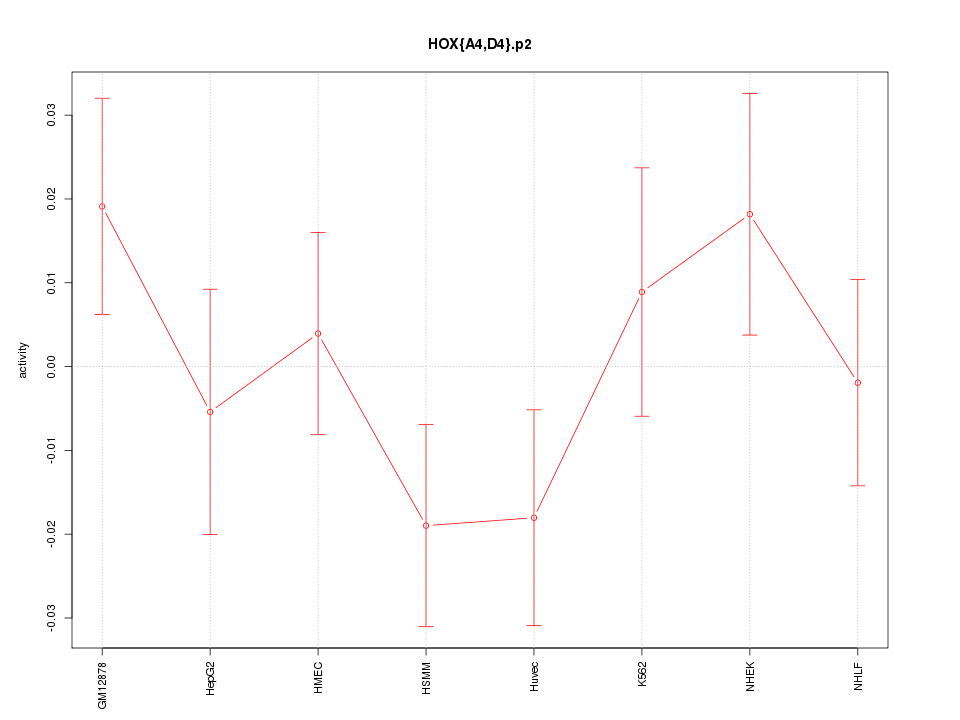
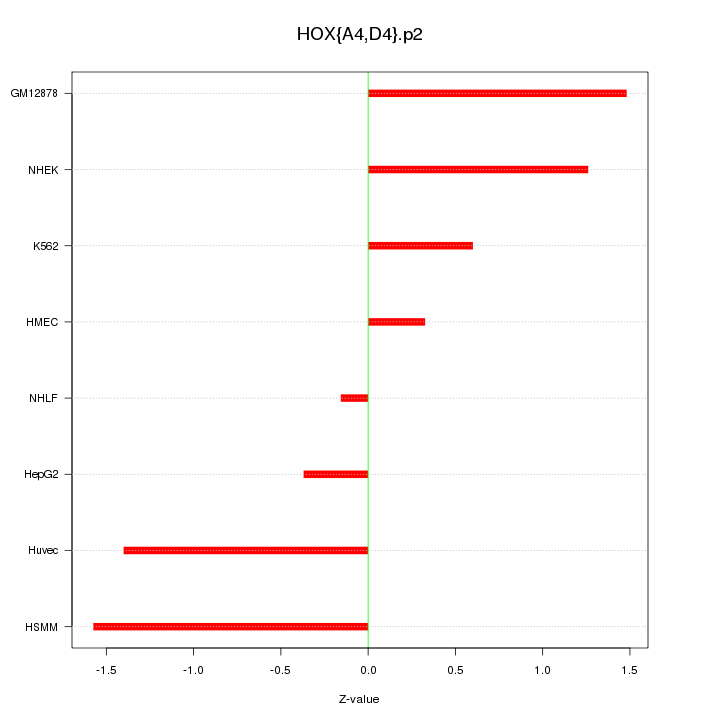
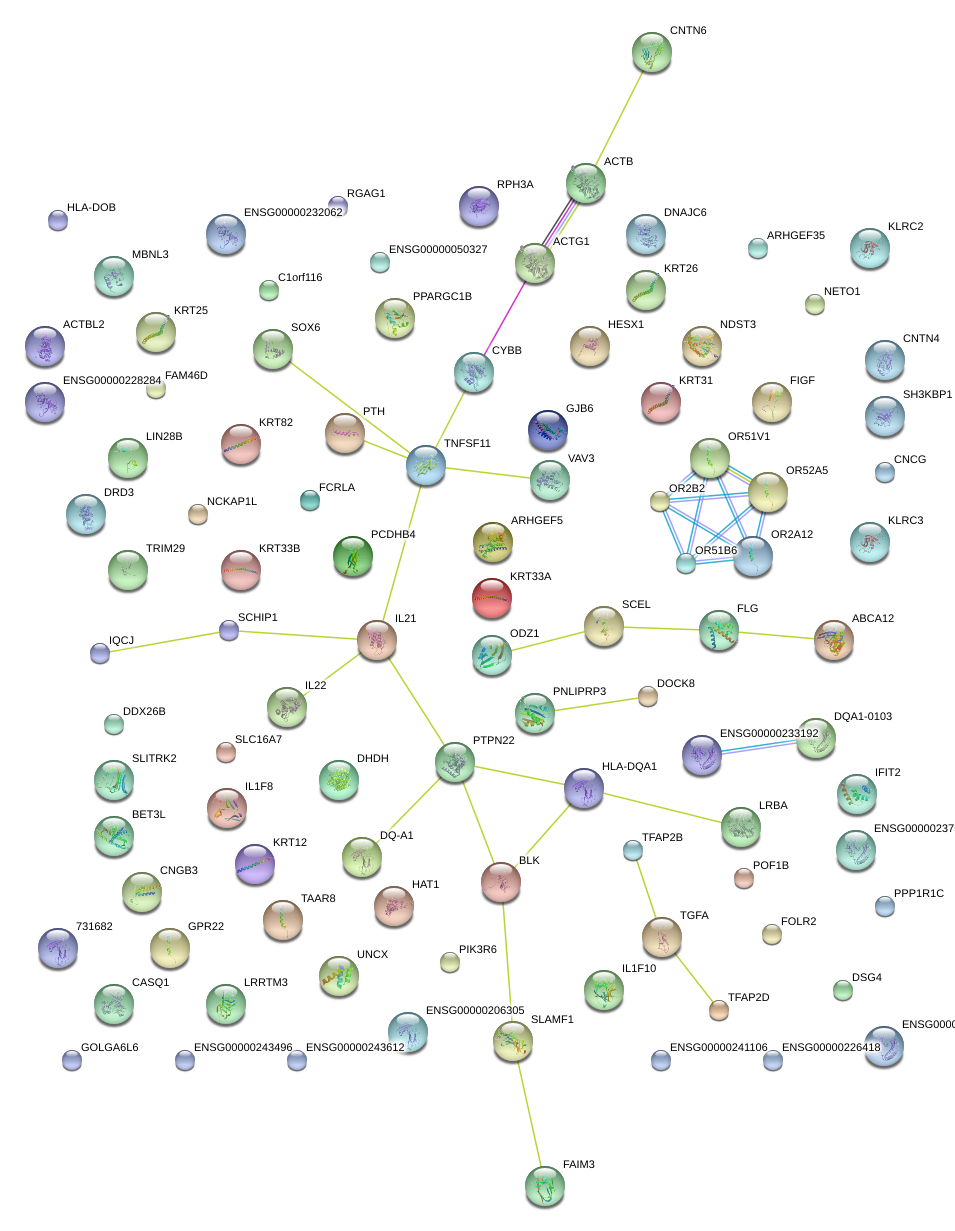
|
chr1_-_205161821
|
1.283
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr11_-_16380967
|
1.056
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_+_5329313
|
0.778
|
NM_001004750
|
OR51B6
|
olfactory receptor, family 51, subfamily B, member 6
|
|
chr6_+_50894397
|
0.763
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr5_-_168204430
|
0.753
|
|
|
|
|
chr2_+_182558795
|
0.696
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr12_+_53177789
|
0.671
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr12_+_53177806
|
0.659
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr3_-_57209319
|
0.658
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr12_+_53177758
|
0.657
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_-_36181927
|
0.629
|
NM_181539
|
KRT26
|
keratin 26
|
|
chr3_+_182900081
|
0.624
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr9_+_263025
|
0.624
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr6_-_32892705
|
0.622
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr12_+_58369392
|
0.620
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr13_-_19703371
|
0.601
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr11_-_5178505
|
0.598
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr1_-_114215768
|
0.596
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr7_+_143423073
|
0.592
|
NM_001004135
|
OR2A12
|
olfactory receptor, family 2, subfamily A, member 12
|
|
chr11_+_71605466
|
0.574
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr5_+_149131695
|
0.566
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_108032645
|
0.557
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_115380539
|
0.548
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chrX_+_79562601
|
0.527
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chrX_+_134482212
|
0.526
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chrX_-_19598912
|
0.523
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_+_53178834
|
0.518
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_-_36165050
|
0.512
|
NM_181534
|
KRT25
|
keratin 25
|
|
chrX_+_37524250
|
0.507
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chrX_-_123925343
|
0.502
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1(Drosophila)
|
|
chr12_-_10464308
|
0.491
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr6_-_27988152
|
0.489
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr7_+_1239078
|
0.483
|
NM_001080461
|
UNCX
|
UNC homeobox
|
|
chr7_+_143683365
|
0.468
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr10_+_68355797
|
0.467
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr1_-_205272619
|
0.455
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr13_+_77007809
|
0.452
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr22_+_21065134
|
0.448
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chrX_-_84521398
|
0.446
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chrX_+_144710557
|
0.439
|
NM_001144006
NM_001144008
NM_001144009
NM_001144010
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr17_-_36779399
|
0.436
|
NM_002279
|
KRT33B
|
keratin 33B
|
|
chrX_+_37524190
|
0.434
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr6_+_132915522
|
0.434
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr7_-_143523582
|
0.432
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chrX_-_84521320
|
0.422
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr7_-_143622188
|
0.421
|
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr17_-_36276987
|
0.421
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr5_+_140481763
|
0.419
|
NM_018938
|
PCDHB4
|
protocadherin beta 4
|
|
chr4_-_47678187
|
0.419
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr6_-_116973465
|
0.415
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr6_+_50789215
|
0.414
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr11_-_119514058
|
0.414
|
NM_012101
|
TRIM29
|
tripartite motif containing 29
|
|
chr6_+_32713160
|
0.413
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr3_+_1109619
|
0.409
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chrX_+_109548940
|
0.402
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr10_+_118177371
|
0.393
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr5_-_56814310
|
0.388
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr20_-_30703219
|
0.376
|
NM_182584
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr1_+_159943385
|
0.376
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr10_+_91051685
|
0.374
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chrX_+_65152004
|
0.366
|
|
|
|
|
chr12_-_51086330
|
0.361
|
NM_033033
|
KRT82
|
keratin 82
|
|
chr15_-_19007127
|
0.360
|
NM_001145004
|
GOLGA6L6
|
golgin A6 family-like 6
|
|
chr1_+_158426908
|
0.355
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr17_-_8711701
|
0.354
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr12_+_111713931
|
0.352
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr3_+_2528272
|
0.352
|
|
CNTN4
|
contactin 4
|
|
chr1_+_65503011
|
0.350
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr13_+_42034871
|
0.349
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chrX_-_15312390
|
0.344
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr1_-_158883666
|
0.339
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr8_-_87824996
|
0.336
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr18_-_68683913
|
0.336
|
NM_138999
NM_153181
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr8_+_11388918
|
0.335
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr3_+_160269836
|
0.335
|
|
IQCJ
|
IQ motif containing J
|
|
chr19_+_54128750
|
0.335
|
NM_014475
|
DHDH
|
dihydrodiol dehydrogenase (dimeric)
|
|
chr11_-_13474101
|
0.334
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr7_+_106897737
|
0.331
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr12_-_92001543
|
0.327
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr2_+_113542017
|
0.326
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr6_+_105511615
|
0.321
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr2_-_113526910
|
0.317
|
NM_014438
NM_173178
|
IL1F8
|
interleukin 1 family, member 8 (eta)
|
|
chrX_-_131401319
|
0.316
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr4_-_151990061
|
0.315
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr2_-_215605054
|
0.312
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr2_-_70634129
|
0.310
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr11_-_5110383
|
0.310
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chr4_+_119174947
|
0.307
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr2_+_172530120
|
0.306
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr18_+_27210737
|
0.305
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr4_-_123761615
|
0.304
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr19_+_14554895
|
0.302
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr15_+_88545510
|
0.302
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr12_+_47205681
|
0.301
|
NM_001005203
|
OR8S1
|
olfactory receptor, family 8, subfamily S, member 1
|
|
chr11_-_117589283
|
0.301
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr9_+_126460511
|
0.298
|
|
|
|
|
chr12_-_7793335
|
0.297
|
NM_130441
NM_203503
|
CLEC4C
|
C-type lectin domain family 4, member C
|
|
chr4_-_38710376
|
0.295
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chrX_+_134482387
|
0.295
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr12_-_101398429
|
0.294
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr11_+_5466490
|
0.294
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr10_-_69095362
|
0.291
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chrX_-_11194015
|
0.290
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr13_-_83354474
|
0.289
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr1_+_207668787
|
0.288
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr19_-_7672978
|
0.287
|
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr5_-_95044415
|
0.286
|
NM_031952
|
SPATA9
|
spermatogenesis associated 9
|
|
chrX_+_12795111
|
0.284
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr11_+_5925152
|
0.284
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr16_-_56562512
|
0.282
|
NM_001135639
NM_001297
|
CNGB1
|
cyclic nucleotide gated channel beta 1
|
|
chr6_+_154449334
|
0.279
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr15_+_38319326
|
0.279
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr11_+_4466684
|
0.278
|
NM_001005171
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1
|
|
chr2_+_79593679
|
0.277
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr5_-_147142444
|
0.277
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_+_99288706
|
0.277
|
NM_054106
|
OR5AC2
|
olfactory receptor, family 5, subfamily AC, member 2
|
|
chrX_+_99726445
|
0.276
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr11_+_59979857
|
0.275
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr19_+_55965532
|
0.274
|
NM_001506
|
GPR32
|
G protein-coupled receptor 32
|
|
chr5_+_176494690
|
0.271
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr6_+_28032997
|
0.271
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr4_+_71298187
|
0.269
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr2_+_17861266
|
0.265
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr1_+_559618
|
0.263
|
|
|
|
|
chr4_+_71235259
|
0.262
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr2_+_225973845
|
0.262
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr7_+_128293699
|
0.261
|
NM_001195150
|
LOC100130705
|
hypothetical protein LOC100130705
|
|
chr1_+_556968
|
0.260
|
|
|
|
|
chrX_+_92815667
|
0.260
|
NM_001171109
NM_001171110
NM_001171111
NM_173698
|
FAM133A
|
family with sequence similarity 133, member A
|
|
chr18_+_59295123
|
0.259
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr5_-_14706486
|
0.258
|
|
|
|
|
chr12_-_7547552
|
0.258
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr1_+_159959072
|
0.257
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr1_-_203579932
|
0.257
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr1_+_559129
|
0.256
|
|
|
|
|
chr1_+_198263352
|
0.255
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr3_-_74652952
|
0.255
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr10_-_105835609
|
0.254
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr4_+_70928718
|
0.253
|
NM_000200
|
HTN3
|
histatin 3
|
|
chrX_-_137649180
|
0.253
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr17_-_3408038
|
0.253
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr1_-_42738535
|
0.251
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr5_-_147142457
|
0.251
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_-_191322919
|
0.251
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr1_-_150029529
|
0.248
|
NM_001083963
NM_001083964
NM_001083965
NM_006862
|
TDRKH
|
tudor and KH domain containing
|
|
chr12_-_54150188
|
0.247
|
NM_001005499
|
OR6C70
|
olfactory receptor, family 6, subfamily C, member 70
|
|
chr9_+_74326536
|
0.247
|
NM_138691
|
TMC1
|
transmembrane channel-like 1
|
|
chr4_-_64957595
|
0.246
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr4_-_40212670
|
0.246
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr18_+_59705918
|
0.244
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr7_+_116447499
|
0.244
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_-_158815850
|
0.244
|
|
CD84
|
CD84 molecule
|
|
chr21_-_30460841
|
0.244
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr14_-_104602784
|
0.243
|
NM_013345
|
GPR132
|
G protein-coupled receptor 132
|
|
chr1_-_158759606
|
0.243
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr7_+_141457977
|
0.241
|
|
LOC93432
|
maltase-glucoamylase-like pseudogene
|
|
chr5_-_147142249
|
0.241
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_+_938709
|
0.241
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr9_+_92629525
|
0.240
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chr9_+_86474445
|
0.240
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr11_-_119514011
|
0.239
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr13_-_83354425
|
0.238
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr17_+_51585834
|
0.235
|
NM_153228
|
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1
|
|
chr10_+_129237602
|
0.235
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr11_-_5037432
|
0.231
|
NM_001005164
|
OR52E2
|
olfactory receptor, family 52, subfamily E, member 2
|
|
chr14_-_104602809
|
0.231
|
|
GPR132
|
G protein-coupled receptor 132
|
|
chr19_-_47623340
|
0.231
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr1_-_156923111
|
0.229
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr7_+_142590633
|
0.228
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chr7_-_138014329
|
0.228
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr8_+_19840861
|
0.228
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr7_-_143622233
|
0.225
|
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr10_-_123343320
|
0.225
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr4_+_71492581
|
0.223
|
NM_016519
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr1_-_167947301
|
0.223
|
NM_000655
|
SELL
|
selectin L
|
|
chr1_+_40631603
|
0.221
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr12_-_101398470
|
0.221
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_+_69356827
|
0.220
|
NM_001178087
NM_022978
NM_021967
NM_022968
|
SERF1B
SERF1A
|
small EDRK-rich factor 1B (centromeric)
small EDRK-rich factor 1A (telomeric)
|
|
chr19_-_20399601
|
0.220
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr1_-_34404029
|
0.219
|
NM_052896
|
CSMD2
|
CUB and Sushi multiple domains 2
|
|
chr19_-_55560686
|
0.218
|
NM_004851
|
NAPSA
|
napsin A aspartic peptidase
|
|
chr12_-_11030777
|
0.218
|
NM_176890
|
TAS2R50
|
taste receptor, type 2, member 50
|
|
chr14_+_19458580
|
0.218
|
NM_001005483
|
OR4K5
|
olfactory receptor, family 4, subfamily K, member 5
|
|
chr16_+_56286212
|
0.218
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr17_-_36209672
|
0.217
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr4_-_72868598
|
0.216
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr15_-_70838703
|
0.216
|
|
|
|
|
chr12_-_52016270
|
0.216
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr11_-_102250876
|
0.216
|
NM_002426
|
MMP12
|
matrix metallopeptidase 12 (macrophage elastase)
|
|
chr4_-_74704964
|
0.215
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr3_-_18455207
|
0.215
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr7_+_128802719
|
0.215
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_149004852
|
0.214
|
|
CTSS
|
cathepsin S
|
|
chr2_-_158008763
|
0.214
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr11_+_125163215
|
0.214
|
NM_001129883
|
PATE3
|
prostate and testis expressed 3
|
|
chr2_+_113591940
|
0.213
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr14_-_56342097
|
0.212
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr13_-_46368968
|
0.212
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr19_+_17723285
|
0.212
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chrX_-_114158388
|
0.211
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|